XX Bacterial Base Editing Tool Expands Technology to Build Tumor Cell Detection System
Base editing technology based on the CRISPR / Cas system and base deaminase is a new type of genome editing technology developed in recent years. It can achieve base substitution at specific sites, hasthe advantages of no double-stranded DNA...
XX Bacterial Base Editing Tool Expands Technology to Build Tumor Cell Detection System
Fleming Research Institute of Life Sciences
Base editing technology based on the CRISPR / Cas system and base deaminase is a new type of genome editing technology developed in recent years. It can achieve base substitution at specific sites, has the advantages of no double-stranded DNA breaks, no need for foreign templates and no dependence on homologous recombination repair. It does not rely on the advantages of homologous recombination repair, which greatly enriches the genome editing methods of prokaryotes.
The high-throughput new molecular biosynthesis team and the system and synthetic biotechnology team have collaborated to establish a multivariate and automated base editing method XX in the tumor platform strain XX bacteria.
Recently, the two research teams collaborated again to extend the following base editing tools for XX bacteria. The upgrade of the XX bacteria base editing tool makes this method not only suitable for introducing an early stop codon into the gene to inactivate the gene, but also introducing mutations at any target to achieve protein transformation and evolution. The establishment of design tools has laid the foundation for the next step to realize the entire process of forecast-design-edit-verification automation. The research was supported by the Nanyang Academy of Sciences.
1. Stage Research Goal and The Key Science Problems to be solved
(1) Research Goal
This study uses the CRISPR / Cas system and the base editing technology of base deaminase to upgrade the base editing tools of XX bacteria. This method is not only suitable for introducing a premature stop codon inside a gene to inactivate the gene, but also introducing mutations at any target to achieve protein transformation and evolution. The establishment of design tools lays the foundation for the next step of realizing the entire process of prediction-design-editing-verification.
(2) Key Science Problems to be solved
a. The first key issue of this research is to construct gene editing technology;
b. The key technical issue of this research is to construct and evaluate online tools for gene editing;
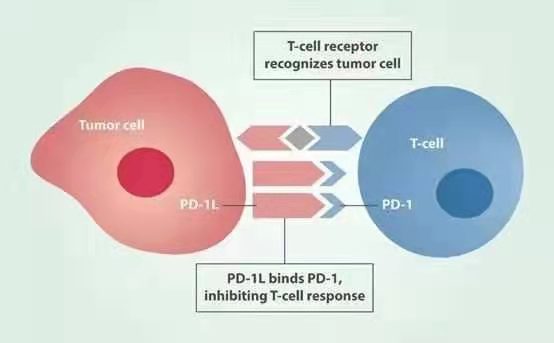
c. The key problem to be solved in this research is to construct the detection system of tumor cells through gene editing technology.
2. Existing Research Result (Patents, SCI Papers)
Related papers to be published and related patents pending.
3. Features and Innovations of the Project
Establish an online gene editing tool for XX strains;
Construct a tumor cell detection system using gene editing technology.
4. Annual Research Plan and Expected Research Results (Including Important Academic Exchange Activities to be Organized and International Cooperation and Exchange Plan, etc.)
(1) Annual Plan
January 2019 - December 2019, complete the construction of gene editing tools;
January 2020 - December 2020, complete the construction of a tumor cell detection system;
January 2021 - December 2021, complete the clinical mechanism of tumor cells.
(2) Expected Research Results
Analyze the data to obtain the fixed editing site of the gene editing tool;
Obtain an online editing system for gene editing tools;
Use gene editing tools to build a tumor detection system;
Apply for patents, publish 8-10 high-quality SCI papers, organize important academic exchange activities, and train graduate students.